Difference between revisions of "SICdb Documentation"
| (6 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
[[File:Logo1HD.png|frameless|center]] | |||
== Introduction == | == Introduction == | ||
{{:Introduction}} | {{:Introduction}} | ||
== Dataset, Database and Software == | === About Dataset, Database and Software === | ||
The dataset is hosted on PhysioNet. You need to be a credentialed user and sign a data use agreement. Please handle data with care! | The dataset is hosted on PhysioNet. You need to be a credentialed user and sign a data use agreement. Please handle data with care! | ||
| Line 27: | Line 32: | ||
=== Data Description medication.csv.gz === | === Data Description medication.csv.gz === | ||
{{:Data Description Medication}} | {{:Data Description Medication}} | ||
=== Data Description | === Data Description data_range.csv.gz === | ||
{{: | {{:Data_Description_Data_Range}} | ||
=== Data Description unitlog.csv.gz === | |||
{{:Data Description Unitlog}} | |||
== Preprocessed fields == | == Preprocessed fields == | ||
Latest revision as of 10:35, 9 November 2022
Introduction
The SICdb dataset provides insight to over 27 thousand intensive care admissions, including therapy and data of their preceding surgery. Data was collected between 2013 and 2021 from 4 of the intensive care units at the University Hospital Salzburg, having more than 3 thousand intensive care admissions per year on 41 beds. The dataset is deidentified and contains, amongst others, case information, laboratory, medication, monitor and respirator signal data.SICdb provides aggregated once-per-hour and highly granular once-per-minute data.
The dataset, version 1.0.8, includes data from more than 27,350 admissions to the Department of Anesthesiology and Intensive Care Medicine at the General Hospital Salzburg and Paracelsus Medical University.
Data Tables
The SICdb dataset consists of billions of data entries across 7 data tables. The main table, "cases," contains a single entry for each intensive care admission and includes information about the patient (such as age, weight, and sex) and case details (such as diagnosis, scores, and ICD10 codes). The "TimeOfStay" field indicates the time from the first admission to a Metavision-enabled ward to the final closing of the case, including any preceding surgery. The "OffsetOfDeath" field indicates survival in seconds from admission. This data represents in-hospital mortality and may extend beyond the length of the intensive care stay or the current hospital stay. Personal data such as age, weight, and height have been grouped into bins of 5, with ages over 90 placed in the final bin.
All other data tables are related to the "cases" table through the "CaseID" field. Most data has timing information, with the "offset" field indicating the number of seconds from admission to the time of the event. The "laboratory" table contains laboratory values and the "medication" table provides data on administered drugs. There are several generic data tables that contain data sorted by type. The "data_ref" table contains additional nominal/categorical data, one entry per admission, and the "data_range" table documents items with a start and end time, such as data on central lines or drainages. The "data_float_h" table contains float data, aggregated once per hour, and includes most signal data. To reduce table size, minute data is serialized as a stream of IEEE 754 floats in the "data_float_h.rawdata" field. For further instructions on using this data, see the documentation found in Documentation.pdf or online [7], and refer to the unpacking script example provided on our GitHub repository [8].
Reference Table
Nominal data is encoded, and the reference table "d_references" provides additional information about the associated field. The referenced fields in all data tables correspond to the primary key of the "d_references" table, "ReferenceGlobalID." The "ReferenceValue" field in "d_references" gives the variable's value, and the "ReferenceUnit" field holds the unit or measurement, if applicable.
Data Format
GZip-compressed RFC 4180 comma-separated files are provided. The most current documentation, including table schemas, can be found online , and an offline copy is included in the files under the name Documentation.pdf. A GitHub repository has been created to share code, report issues, and discuss the dataset.
About Dataset, Database and Software
The dataset is hosted on PhysioNet. You need to be a credentialed user and sign a data use agreement. Please handle data with care!
We provide a software package to build up da database, select and export data into various file formats. Refer to https://www.sicdb.com/Documentation/Get_Started to start!
File Description
The SICdb dataset consists of 8 files. All data files in the SICdb dataset are provided as comma seperated files (.csv) using the RFC 4180 standard. Additionally all files are compressed with gzip, an commonly used openly available compression method.
List of files
Data Description cases.csv.gz
"The "cases" table is the base table of the SICdb dataset and the "CaseID" field serves as the identifier that relates all data. Each admission to the intensive care unit generates a unique "CaseID." To identify readmissions, a "PatientID" is provided. The "OffsetAfterFirstAdmission" field stores the time between the first admission and the current one.
"TimeOfStay" is the time in seconds from the admission recorded by the MetaVision system to the discharge. This time may include any preceding surgery and ends when the case is closed.
There are three fields indicating survival: "DischargeState" indicates the patient's state upon discharge from the ICU and is taken from the MetaVision discharge form. "HospitalDischargeType" provides information on how the case was closed in the clinical information system. "OffsetOfDeath" is the time in seconds from admission to death and includes additional hospital stays and out of hospital mortality. As it refers to a 1-year mortality it is set to null if death occurred more than one year after admission.
| Name | Type | Description | Comment |
|---|---|---|---|
| CaseID | Integer | A randomly assigned identifier | CaseID is unique to each admission. |
| PatientID | Integer | A randomly assigned identifier | PatientID is unique to each Patient, useful to identify readmission |
| AdmissionYear | Integer | Year of admission | |
| TimeOfStay | Integer | Time of stay (seconds) | Time from primary metavision admission, to last discharge. In cases, where the patient is admitted to ward after surgery, this will include surgery time. |
| ICUOffset | Integer | Time of actual ICU admission | SICdb includes preceding surgery if applicable, this field indicates the first transfer to an intensive or intermediate care ward |
| saps3 | Float | Simplified Acute Physiology Score III | |
| HospitalDischargeType | Reference | Type of hospital discharge. | This field indicates survival. |
| HospitalDischargeDay | Integer | The day of hospital discharge after admission | Note that this data is only available in days |
| HospitalStayDays | Integer | Days stayed in hospital for this case, including pre icu. | |
| DischargeState | Reference | Type of icu discharge | This field indicates survival on icu |
| DischargeUnit | Reference | Unit the patient was discharged to, as selected in Metavision discharge form | |
| OffsetOfDeath | Integer | 1-year mortality in seconds from primary admission to death | 1 year mortality, including out-of-hospital as far as known. See The "OffsetOfDeath" field denotes the elapsed time, in seconds, from admission to death, taking into account any subsequent hospital stays and out-of-hospital mortality data. The data was gathered from various sources, including government data. It is set to null if the death occurs more than one year after admission, since it is defined to 1-year mortality. For technical reasons observation time is only 6 month for some patients, field "EstimatedSurvivalObservationTime" holds information about that. However, it is worth noting that in some cases, if a patient dies in foreign countries, the information may be scarce. Postal address validity checks were performed, but no statistically significant differences were found, indicating that this may not be an issue of concern. for further information. |
| EstimatedSurvivalObservationTime | Reference | Estimation of oversation time, either 1-year or 6-month | |
| Sex | Reference | ||
| WeightOnAdmission | Float | Rounded to +-5kg | |
| HeightOnAdmission | Float | Rounded to +-5cm | |
| AgeOnAdmission | Integer | Rounded to +-5y, over 90 set to 90 | |
| HospitalUnit | Reference | Last unit using this case | |
| ReferringUnit | Reference | Referring unit as selected in admission form | Note: Unfortunatly in some cases "Notaufnahme" is selected here, so the referring unit is not specified in these cases. |
| ICD10Main | Text | ICD10 main code | |
| ICD10MainText | Text | ICD10 main text | |
| SurgicalSite | Reference | ||
| InterventionsText | Text | List of interventions | |
| HoursOfCRRT | Integer | Hours of continuous renal replacement threapy this admission ** | |
| AdmissionFormHasSepsis | Reference | A mandatory field in the admission form ** | |
| HeartSurgeryAdditionalData | For heart surgery patients there is additional data collected ** | Yes if applicable | |
| HeartSurgeryCPBTime | Integer | Bypass time ** | |
| HeartSurgeryBeginOffset | Integer | Offset in seconds from ICU admission to cut ** | |
| HeartSurgeryEndOffset | Integer | Offset in seconds from ICU admission to end of surgery** | |
| OffsetAfterFirstAdmission | Integer | If a patient has more than one admission, this is the offset in seconds from the first |
* These fields are not available on PhysioNet at the moment. Contact us for further information. ** These fields will be moved to data tables in version 1.1.0
Data Description d_references.csv.gz
The d_references table contains information on all encoded data fields of the SICdb dataset. Each field, that has "Reference" as field type, is associated with the ReferenceGlobalID in the d_references table. Additionally ReferenceUnit describes the unit of measurement used for this field. Refer to chapter SQL Examples to learn how to easily use this table in relational databases.
| Name | Type | Description | Comment |
|---|---|---|---|
| ReferenceGlobalID | Integer | The unique ID for the reference | Use this identifier as dictionary for alle encoded fields |
| ReferenceValue | Text | Reference value | i.e. "Creatinine" |
| ReferenceName | Text | The name of the reference | i.e. "Laboratory" |
| ReferenceUnit | Text | The unit of this item if applicable | i.e. "mg/dl" |
| LOINC_code | Text | LOINC Code | |
| LOINC_short | Text | LOINC SHORTNAME | |
| LOINC_long | Text | LOINC LONG_COMMON_NAME |
Data Description data_float_h.csv.gz
Contains hourly float data associated with a case. If more (minute) data is available, data was aggregated and the corresponding values are stored in rawdata field. Refer to Deserialize Raw Data for more information. Metavision does, in database, not differentiate between qualitative (i.e. blood pressure) and quantitative (i.e. drainage volume) data, this was done programatically saving the sum instead of average. The field cnt indicates the amount of values aggregated.
| Name | Type | Description | Comment |
|---|---|---|---|
| CaseID | Integer | Case identifier | |
| DataID | Reference | ||
| Offset | Integer | Time in seconds after admission | |
| Val | Float | Value | The unit (if applicable) is found in d_references where data_float_h.DataID is associated to d_references.ReferenceGlobalID |
| cnt | Integer | Amount of values aggregated | |
| rawdata | Blob | List of 60 floats containing raw data | Due to excessive storage needs minute values have been aggregated. |
Refer to chapter "Raw Data" for more information
Data Description data_ref.csv.gz
data_ref.csv.gz contains referenced (nominal) data and is unique per icu admission.
| Name | Type | Description | Comment |
|---|---|---|---|
| id | Integer | Primary Key | |
| CaseID | Integer | Case identifier | |
| FieldID | Reference | Refers to the name of the field | i.e. PreconditionDiabetes |
| RefID | Reference |
Data Description laboratory.csv.gz
The laboratory table contains all available lab data, in many cases including the pre-admission labs.
Like all "Reference" fields additional data like name and unit of measurement is found in d_references. See SQL Examples to learn how to connect them using a relational database system.
To simplify lookup for labs of previous stays PatientID was added to this table.
| Name | Type | Description | Comment |
|---|---|---|---|
| id | Integer | Primary Key | |
| CaseID | Integer | Case identifier | |
| PatientID | Integer | Patient identifier | |
| DrugID | Reference | ||
| Offset | Integer | Time in seconds after admission | |
| LaboratoryValue | Float | Value | Unity of measurement can be found in d_references |
| LaboratoryType | Reference | Used for special laboratory types, i.e. arterial blood gas. |
Data Description medication.csv.gz
Contains all medication given during stay. Additional data like name and unit of measurement is found in d_references, where medication.DrugID is associated to d_references.ReferenceGlobalID. Metavision does not differ between continuous or single doses in rangesignals, so a single dose is set to 60 second admission time. Each change of a continuous application will end the current entry and create a new one.
| Name | Type | Description | Comment |
|---|---|---|---|
| id | Integer | Primary Key | |
| CaseID | Integer | Case identifier | |
| DrugID | Reference | ||
| Offset | Integer | Time in seconds after admission | |
| OffsetDrugEnd | Integer | If drug was given continuously refers to end of application. Field is set to 60 seconds on bolus application. | |
| IsSingleDose | Integer | Set to 1 when given as bolus. | |
| Amount | Float | Full amount given regardless of time | Unity of measurement found in d_references |
| AmountPerMinute | Float | Dosage per minute | Unity of measurement found in d_references |
Data Description data_range.csv.gz
This table contains, amongst others, data about treatment or invasivities like central lines. Data in this table has a DataID, which refers to d_references.ReferenceGlobalID, a begin offset and an end offset.
| Name | Type | Description | Comment |
|---|---|---|---|
| id | Integer | Primary Key | |
| CaseID | Integer | Case identifier | |
| DataID | Reference | ||
| Offset | Integer | Time in seconds from admission | |
| OffsetEnd | Integer | Time in seconds from admission | |
| Data | Text | May contain a json object containing additional data (i.e. endotracheal tube size, ...) |
Note: The data field is currently null. It has nevertheless been included for further compatibility, as it is planned to fill this field in next incremental update, which is sheduled March 2023.
Data Description unitlog.csv.gz
The unitlog table contains information of patient transportation. Note that in many cases Metavision creates multiple entries and this data is difficult to interpret.
| Name | Type | Description |
|---|---|---|
| id | Integer | Primary Key |
| CaseID | Integer | |
| PatientID | Integer | |
| LogState | Integer | Internal Metavision value. Note that in this version of the dataset this is a raw unencoded value. |
| Offset | Integer | Time in seconds after admission |
| HospitalUnit | Reference |
Preprocessed fields
Several fields have been preprocessed and added to the database. This includes peprocessed-for-convenience and preprocessed-for-anonymization elements. The data is found in data_ref.csv.gz, the text values are referenced in d_references, where d_references.ReferenceGlobalID relates to data_ref. FieldID and data_ref.RefID.
| Name | Type | Description |
|---|---|---|
| PremedicationBetablocker | Referenced | Yes/No |
| PremedicationACEInhibitor | Referenced | Yes/No |
| PremedicationStatin | Referenced | Yes/No |
| PremedicationDiuretic | Referenced | Yes/No |
| PreconditionDiabetes | Referenced | Any type of diabetes (IDDM or NIDDM) |
| PreconditionLungDisease | Referenced | Any kind of lung severe disease (COPD, restrictive lung diseases, ...). Asthma is included when medication is taken daily |
| PreconditionArtHypertension | Referenced | Any kind of treated or diagnosed arterial hypertension |
| PreconditionRenalDysfunction | Referenced | If stated in admission protocol (omitted due to anonymity issues) |
Case Selection
The SICdb dataset contains all admissions to metavision, that were treated on an intensive care unit. This includes patients that are admitted post-surgery as well as internal medicine cases. Note that non-surgical reasons for admission are highly underrepresentated in this dataset because most internal medicine patients are treated on a unit not using an electric PDMS.
All admissions to participating intensiv care units are included, as long as following conditions do not apply:
- Less the 10 minutes of Stay
- Missing mandatory admission data
- No monitor signals
- No medication data
- hospital number mismatch
Intensive care or intermediate care?
To provide as much data as possible all patients admitted to one of our intensive or intermediate care units are included. While the associated beds have a designated care level, the patients treatment level is not defined and are treated according to their needs, they may be transferred multiply between the strongly cooperating units. In general, patients that are uniquely associated (see unitlog) to INIC or INID did not receive high level intensive care treatment. Please note that the field cases.HospitalUnit gives no information about the level of intensive care a patient received. We may provide a preprocessed field in future versions of the SICdb dataset concerning the level of treatment.
Unit Description
The participating metavision enabled wards are CWIN, INBD, INIC, and INIC. INCV and CWCV are used for cases associated with SARS-COV-2, this includes SARS-COV-2 ARDS and patients with coincidental positive test and other cause of intensive care. CWIN and INBD, INCV and CWCVC are high level intensive care units. INIC and INID are lower level of care and do not provide invasive mechanical ventilation (NIV possible on INID, HFNC on all wards), CRRT or ECMO.
Validation
All cases are cross-matched with a second database. In case of unsuccessful matching, cases were excluded, as these refer to entries for testing and demo.
The dataset is in constant development. Feel free to contact us when you find a significant amount of invalid data.
SQL Examples
While the SICdb dataset is provided as csv raw data, it is designed to be used in a a relational database management system. Refer to Get Started for further information building up the database and corresponding software. Our software includes hundreds of configurable one-click SQL snippets like creatinine on day one/two/..., max/min/average lactate and much more.
Using Referenced Fields
One of the most important tasks is decoding the referenced fields. While our Software does this automatically the database design makes it also quite simple in raw SQL code.
Simple Join Query
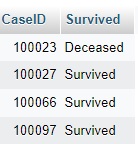
SELECT CaseID, d_references.ReferenceValue as Survived FROM `cases` LEFT JOIN d_references ON d_references.ReferenceGlobalID=cases.HospitalDischargeType;
Join Query including Unit-Of-Measurement
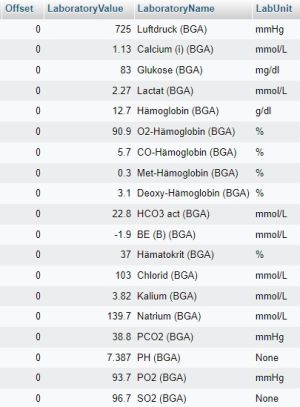
SELECT caseID,Offset,LaboratoryValue,d_references.ReferenceValue as LaboratoryName, d_references.ReferenceUnit as LabUnit FROM `laboratory` LEFT JOIN d_references ON d_references.ReferenceGlobalID=laboratory.LaboratoryID;
Subquery Examples
A quite flexible query solution are subqueries. These allow better configuration for the field. Note this is a MySQL 8.x query and may need to be adapted in other database systems. These SQL code snippets have been generated by our software.
Select Maximum Creatinine Example
SELECT `cases`.`CaseID` AS CaseID ,( SELECT Max(laboratory.LaboratoryValue) FROM laboratory WHERE laboratory.CaseID = cases.CaseID AND laboratory.LaboratoryID = 367 AND Offset >= 0 AND Offset < 7 * 86400 ) AS MaximumCreatinineInFirstWeek FROM cases
Select First Creatinine Example
SELECT `cases`.`CaseID` AS CaseID ,( SELECT laboratory.LaboratoryValue AS val FROM laboratory WHERE laboratory.CaseID = cases.CaseID AND laboratory.LaboratoryID = 367 AND Offset >= 0 ORDER BY laboratory.Offset ASC LIMIT 0 ,1 ) AS FirstCreatinineAfterAdmission FROM cases
Select Second Creatinine Example
SELECT `cases`.`CaseID` AS CaseID ,( SELECT laboratory.LaboratoryValue AS val FROM laboratory WHERE laboratory.CaseID = cases.CaseID AND laboratory.LaboratoryID = 367 AND Offset >= 0 ORDER BY laboratory.Offset ASC LIMIT 1 ,1 ) AS SecondCreatinineAfterAdmission FROM cases
Select Second Creatine With Time Example
A little more advanced example including the time-of-measurement.
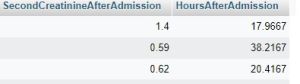
SELECT `cases`.`CaseID` AS CaseID ,( SELECT laboratory.LaboratoryValue AS val FROM laboratory WHERE laboratory.CaseID = cases.CaseID AND laboratory.LaboratoryID = 367 AND Offset >= 0 ORDER BY laboratory.Offset ASC LIMIT 1 ,1 ) AS SecondCreatinineAfterAdmission ,( SELECT laboratory.Offset AS val FROM laboratory WHERE laboratory.CaseID = cases.CaseID AND laboratory.LaboratoryID = 367 AND Offset >= 0 ORDER BY laboratory.Offset ASC LIMIT 1 ,1 ) / 3600 AS HoursAfterAdmission FROM cases
Raw data
Why is this data serialized?
The intermediate data, where minute values are not aggregated, has roughly about 240gb, and it is to be expected that is increases by about 15% in 2023 update. By aggregating and serializing these values the database can be compresssed to under 10gb. Nevertheless it complicates the deserialization of this data. While our software can export the minute values one-click, using it in raw data requires a bit of coding.
Refer to chapter Full Unpack to learn how to completely unpack minute data.
Encoding
The raw data field is a stream of 60 little endian IEEE 754 floats, so it has exactly 240 bytes. The first 4 bytes represent the first minute of the hour and so on. Note that 0x000000 is defined to be NULL (no value).
Python example
def GetRawValues(data):
ret=[]
for i in range(int(len(data)/4)):
if (data[i*4]==0 and data[i*4+1]==0 and data[i*4+2]==0 and data[i*4+3]==0): continue # remove null values
ret.append(struct.unpack('<f',data[i*4:i*4+4])[0])
return ret
This simple example does not include a correct offset for the values. You may check our unpack script to find an example on how to compute the offset.
C# example
public static float?[] GetRawValues(byte[] data)
{
byte[] buf = new byte[4];
float?[] ret = new float?[data.Length / 4];
for(int i = 0; i < data.Length; i += 4)
{
buf[0] = data[i];
buf[1] = data[i+1];
buf[2] = data[i+2];
buf[3] = data[i+3];
if (buf[0] == 0 && buf[1] == 0 && buf[2] == 0 && buf[3] == 0) continue; // ignore null values
ret[i / 4] = BitConverter.ToSingle(buf); // note: if you are on a BigEndian machine you need to flip buf
}
return ret;
}
Full Unpack
We provide a simple unpack python script on our github code repository.
Version Information
The SICdb dataset uses a versioning system. It contains 3 numbers major, minor and patch. A suffix may be added for changes, the do not necessarily needs to be applied.
A major version change generally occurs when the database was incrementally updated and more cases are included.
A minor version change occurs when data was altered that will likely change most study data. (i.e. removal of a significant amount of invalid cases) A minor update also applies when new data is added (i.e. a new table) but the existing data is not altered.
A patch version change occurs when only specific data was altered and will not affect all exports. Please read changelog to check if your data is affected.
1.0.8 (07/2024)
- Added LOINC codes to laboratory references
1.0.7 (04/2024)
- Added field `cases`.`HospitalDischargeDay` `HospitalStayDays` representing the day of release from hospital after admission and the full lenght of hospital stay, respectively.
- Added field `cases`.`AdmissionUrgency`, depicting the urgency of admission
- Added High Flow (HFNC) therapy data
- Added Richmond Agitation-Sedation Scale (RASS) score
- Added Numeric Rating Scale (NRS-11)
- Added SOFA Score
- Removed 36 invalid cases
- Recalculated field `cases`.`OffsetAfterFirstAdmission`, fixing an issue that occasionally led to inadequate values
1.0.6 (05/2023)
- [Important] Update a major error at the heart rate signal (invalid mapping of ecg signal)
- [Important] Renamed heart rate signals
- [Important] Updated some (~150) invalid weight/height signals
- [Fix] Due to a change in table structure the Premedication_ fields missed reference id, fixed
- Added KDIGO_AKI_168 and published algorithm (note: in general be careful with using urine output KDIGO in automated datasets)
- Added field ICUOffset for better comparability with other ICU datasets
- Added more signal data
- Corrected a small issue in the norepinephrine per kg algorithm (change is not significant)
1.0.5
- Merged additional mortality data and changed OffsetOfDeath from in-hospital-mortality to general 1-year-mortality
- Removed or fixed 190 cases of inplausible height or weight data
- Added some more CRRT data
- Recalculated DrainageSum
1.0.4
- Added unity of measurements for signal data
- Structural changes